AnyVLM Features
Variant-Level Matching (VLM) Protocol
AnyVLM implements the nascent Variant-Level Matching, or VLM, protocol. This framework enables construction of a federated genomic knowledge network, allowing laboratories and consortia to make internal variation data discoverable and reusable by other researchers and clinicians. AnyVLM is designed as a lightweight and portable solution for rapidly spinning up a new VLM node and introducing new data to the network.
The VLM protocol is still in development, and is subject to change.
Cohort Allele Frequency (CAF) Retrieval
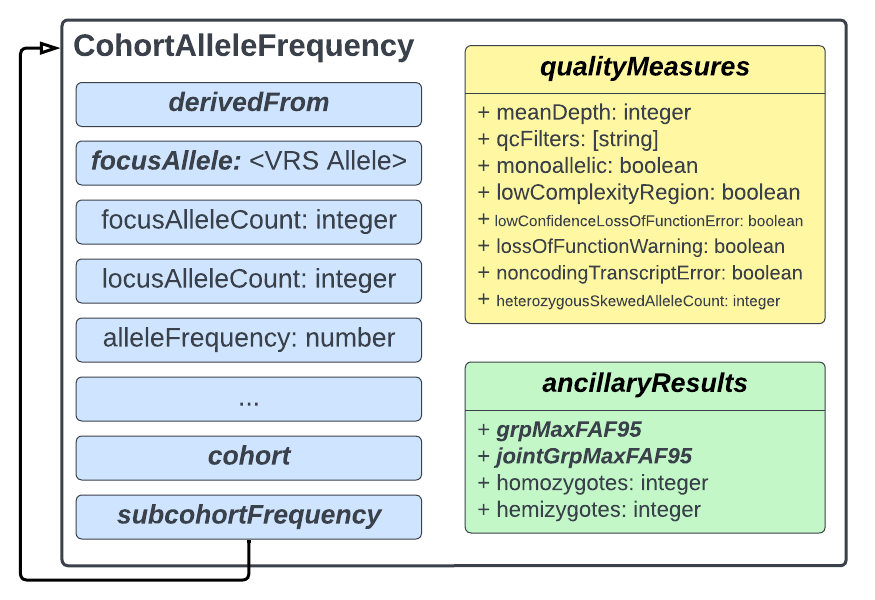
At present, AnyVLM serves Cohort Allele Frequency (CAF) objects, describing the frequency of an allele in a cohort. CAF objects may also report additional data, including frequency broken down by zygosity and quality control filters declared by the original variant calls.
VCF Ingestion
Data is submitted to AnyVLM by way of Variant Call Format (VCF) files. Presently, incoming VCFs must use INFO fields to declare cohort frequency data. See the specific requirements described in the Usage section for more information.
AnyVar Storage Backend
While an AnyVLM instance stores allele frequency data internally, it utilizes AnyVar for registration and retrieval of the variants themselves. An AnyVar instance may be constructed internally, or an external AnyVar instance available on the local network may be used. See the AnyVar client configuration page for information on AnyVar client construction.
Genomics Standards Conformance
This version of AnyVLM validates data using the following data standards:
Standard |
Version |
Datatypes used |
2.0.1 |
||
1.0.1 |